
The CytoSure® Medical Research Exome Array is a highly targeted exon-focused array capable of detecting medically relevant microdeletions and microduplications. This array has been developed in collaboration with leading molecular genetics experts at Emory University and makes an ideal complement to an exome sequencing approach, providing a comprehensive mutation spectrum analysis in rare disease.

Detect single or multiple exonic CNVs

Over 4600 hand-curated, research-validated genes

Select from our proprietary Medical Research Exome database

For confident analysis and reporting
Using a proprietary probe design algorithm, we are able to design highly targeted, optimised probes throughout the majority of the genome. A range of probe performance metrics are evaluated to ensure optimal array performance. A poorly performing probe can result in inaccurate data and even false calls. All of our probes are first tested in silico and scored on quality. Probes with the highest score are printed on an array and tested in the laboratory. These probes are ranked on performance, and only the most accurate, best performing probes are used in the final design. Probes have been selected to target the exonic regions of 4,645 genes. For the majority of genes there are a minimum of 4 probes per exon. For very large exons, probes are distributed evenly along the exon with 1 probe every 125bp. For any array design, it is also important to provide good backbone coverage to ensure accurate normalisation. In the untargeted backbone, the CytoSure Medical Research Exome array has one probe every 42kb. Using optimised probes enables the detection of small amplifications and deletions (Figure 1).
*Data kindly provided by Madhuri Hegde, Ph.D., FACMG, Emory University.
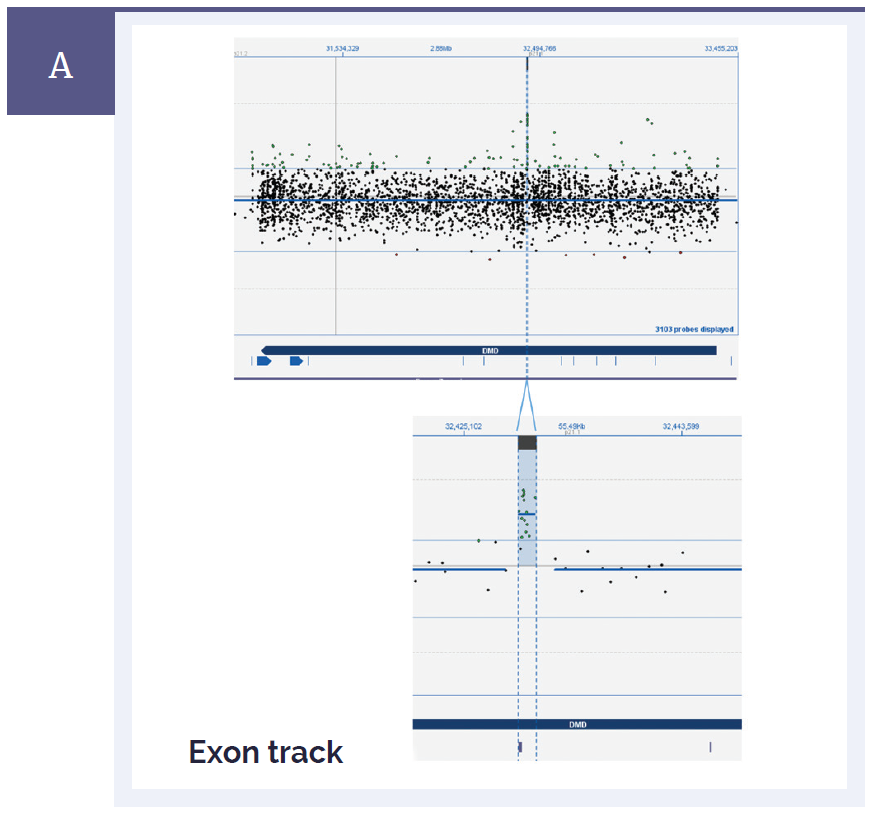
Figure 1a: CytoSure Interpret Software clearly displays small aberrations and enables easy identification of genes and exons. [A] Duchenne muscular dystrophy (DMD) is caused by a mutation in the DMD gene which is one of the largest genes in the genome with over 79 exons. Shown here in the top panel is the complete view of the whole gene. The bottom panel is a close-up view showing a small duplication of 1.4kb in the DMD gene which spans a single exon*.

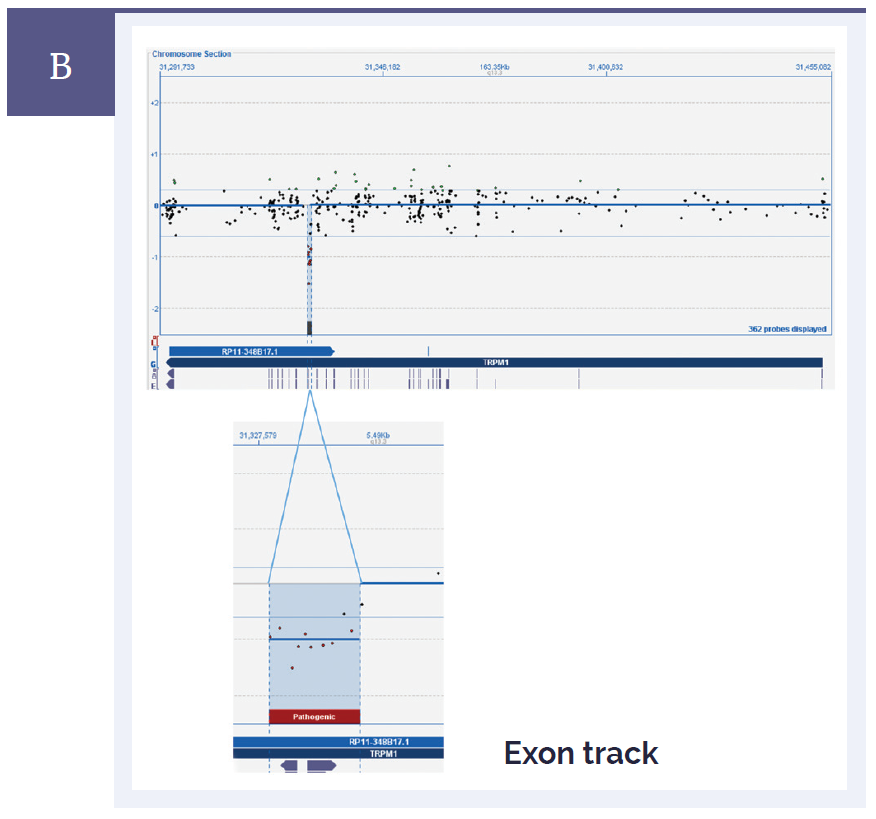
Figure 1b: CytoSure Interpret Software clearly displays small aberrations and enables easy identification of genes and exons. [B] Mutations in TRPM1 are often associated with congenital stationary night blindness. Shown here in the top panel is an overview of the whole gene. In the bottom panel, the close-up view shows a very small 684bp deletion which contains 10 probes, and also spans a single exon*.

Working with the leading clinical research experts at Emory University has enabled us to design an array covering the most relevant regions of the genome gathered from their research into molecular disorders. It has been hand-curated and grouped into disease- and syndrome specific research panels. The content of the CytoSure Medical Research Exome Array is grouped into over sixty different panels. Each panel contains genes relevant to a specific syndrome or genetically linked disease. Working with these panels has enabled Emory Genetics to create a set of ten 4x180k custom arrays containing probes validated on the CytoSure Medical Research Exome Array. These custom arrays provide a more cost-effective, routine option for detailed research into small, exon-specific aberrations.
Genetic research into molecular disorder samples is typically done in batches to ensure cost-effective processing. However, collecting sufficient samples, especially for rare disorders can significantly delay analysis. Combining content for rare disorders with more commonly occurring conditions on to a single array allows immediate processing of a diverse range of samples, thereby reducing both time to analysis and cost. Select from our proprietary database of over 26 million in silico validated probes and build a bespoke custom array based on your laboratory's requirements. Four array formats are available, 8x60k, 4x180k, 2x400k and 1x1M, allowing you to choose the one most suited to your content and throughput needs. Like all of our arrays, custom arrays are optimised to work with our range of high-quality labelling kits and are supplied with CytoSure Interpret Software, our class-leading data analysis package. It is easy to get started as full on-site wet lab and data analysis training is provided as standard. OGT validation data and customer feedback confirms that using CytoSure Genomic DNA Labelling Kits improves DLRS values and signal-to-noise ratios, ensuring accurate detection of even the smallest aberration. Poor quality metrics from using non-optimised kits can indicate an increased level of technical noise on the array which may have an impact on the ability to detect small aberrations.
CytoSure Interpret Software is a powerful, easy-to-use package for the analysis of aCGH data. Innovative features such as the Accelerate Workflow enable standardised and automated data analysis, including automatic aberration detection and classification. It includes extensive annotation tracks covering syndromes, genes, exons, CNVs and recombination hotspots — each of which link to publicly available databases such as ISCA, Decipher and the Database of Genomic Variants and the Cancer Gene Consensus Genes (Figure 3) providing results in context. It is possible to select which tracks are displayed allowing only tracks of specific interest to be viewed (e.g. syndrome-specific tracks) ensuring easy data interpretation. Each track can reference hg18, hg19 or GRCh38 (hg20) information. Annotations within a track can be coloured allowing easy visualisation. It is also possible to customise what information from the tracks is saved in the report (e.g. how many common variants overlap with an aberration).
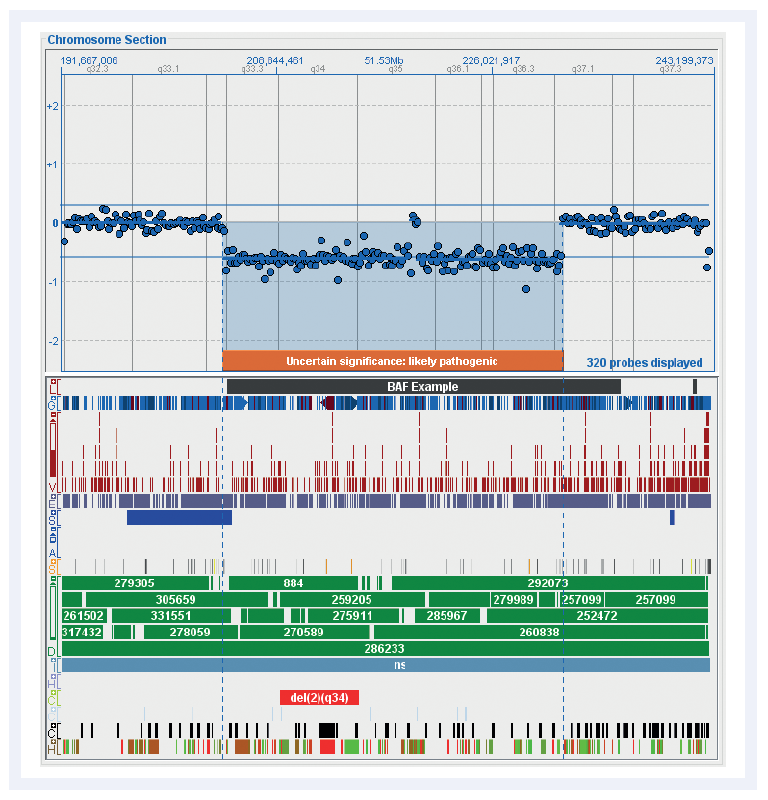
Figure 3: Chromosome overviews are clearly displayed in CytoSure Interpret Software. Shown here is an overview of the distribution of probes on chromosome 2 in the CytoSure Medical Research Exome array and the currently available standard tracks. These fully customisable tracks simplify the interpretation of aberrations.



The OGT MREA provides us with the exon-level resolution needed to detect CNVs over the medical exome that are missed by NGS and traditional microarray designs, providing additional insights into the mutation spectrum of the sample.

Dr. Emily Farrow, Ph.D. CGC
Director of Laboratory Operations, Center for Pediatric Genomic Medicine, Children's Mercy Hospital in Kansas City, USA



