Want to download this as a PDF? Download now
Graham Speight, Giulia Poloni, Natalie Milner, Venu Pullabhatla, James Reid, Jolyon Holdstock, Lyudmila Georgieva (Oxford Gene Technology)
Chronic lymphocytic leukaemia (CLL) is a chronic lymphoproliferative disorder characterised by monoclonal B cell proliferation. It is the most common adult leukaemia in Western populations and comprises 25 to 30% of leukaemias in the United States.
A wide variety of genomic variations are associated with CLL, ranging from single nucleotide variants (SNVs) and insertions/deletions (indels) to complex chromosomal rearrangements such as copy number alterations (CNAs) of part or whole chromosome. Chromosomal abnormalities are very common in CLL, the most frequent being del(13q), trisomy 12, del(11q), del(17p) and del(6q).
Comprehensive genetic research into CLL requires multiple testing strategies with high associated costs. In this study, we tested the capability of OGT’s NGS panel, SureSeq™ CLL + CNV V3, to detect SNVs and indels as well as chromosomal aberrations in CLL research samples, allowing for the detection of CLL variants in a cost efficient and timely manner.
The SureSeq hybridisation-based enrichment approach and OGT’s Universal Complete NGS Workflow Solution were used throughout this study (Figure 1).
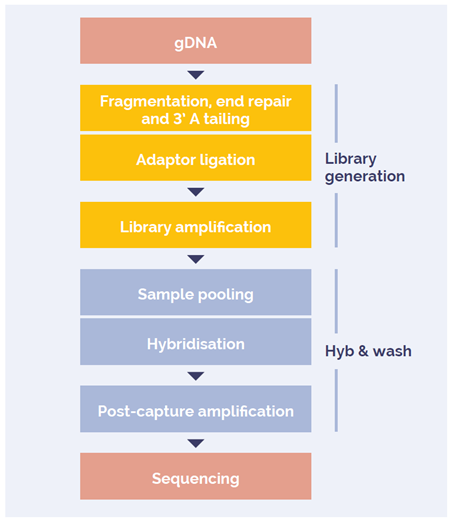 Figure 1: Universal NGS Workflow DNA to sequencer in 1.5 days with minimal handling time.
Figure 1: Universal NGS Workflow DNA to sequencer in 1.5 days with minimal handling time.
The SureSeq CLL + CNV V3 Panel can be used for detection of SNVs and indels in 16 genes and somatic CNAs in 5 critical regions.
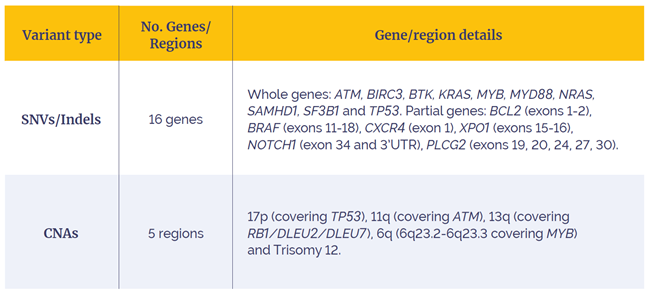 Table 1: SureSeq CLL + CNV V3 Panel content.
Table 1: SureSeq CLL + CNV V3 Panel content.
For each set of 8 samples, one Reference DNA (negative for aberrations in the critical regions) was hybridised and sequenced with the research samples. Information from reference DNA samples are used in the analysis for CNAs detection.
Sequencing was conducted using 2 x 150 bp reads on an Illumina MiSeq® V2 300.
The workflow and the panel were tested on reference DNA (Coriell Institute, Horizon) to validate the overall performance. CNAs capability was demonstrated by using 21 research samples with known CNAs in the regions of interest previously characterised by FISH.
To mimic CNAs at low tumour content, 11 research samples with known aberrations were diluted in order to create CNA carriers with 20-40% tumour content.
Sequencing data analysis including CNA detection was performed using OGT’s Interpret NGS Analysis Software.
High depth and uniform of coverage was achieved for all targeted genes and genomic regions allowing for confident detection of low frequency gene specific SNVs and indels.
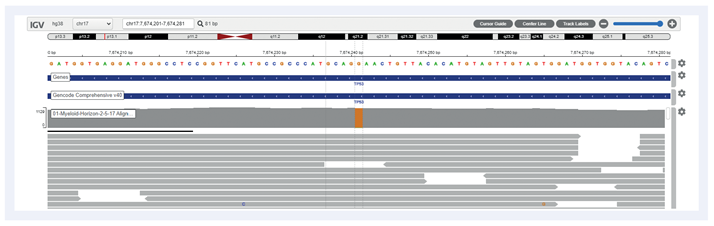 Figure 2: Example of TP53 c.722C>T estimated at 1.62% VAF.
Figure 2: Example of TP53 c.722C>T estimated at 1.62% VAF.
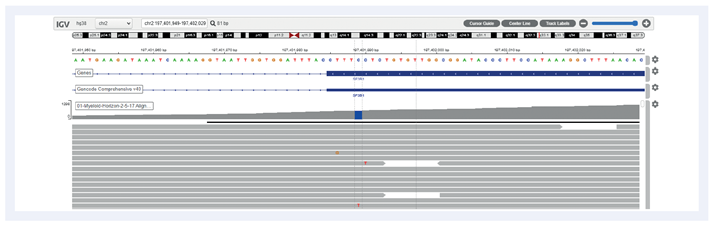
Figure 3: Example of SF3B1 c.2219G>A estimated at 1.04% VAF.
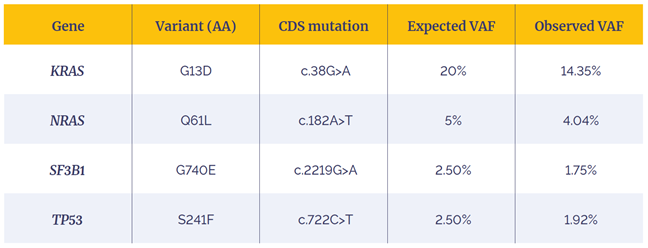
Table 2: Variants present in the Myeloid DNA Reference Standard (Horizon) and detected by the SureSeq CLL + CNV V3 Panel.
Data presented here are from 21 research samples that were processed using the OGT workflow in combination with OGT’s Interpret NGS Analysis Software.
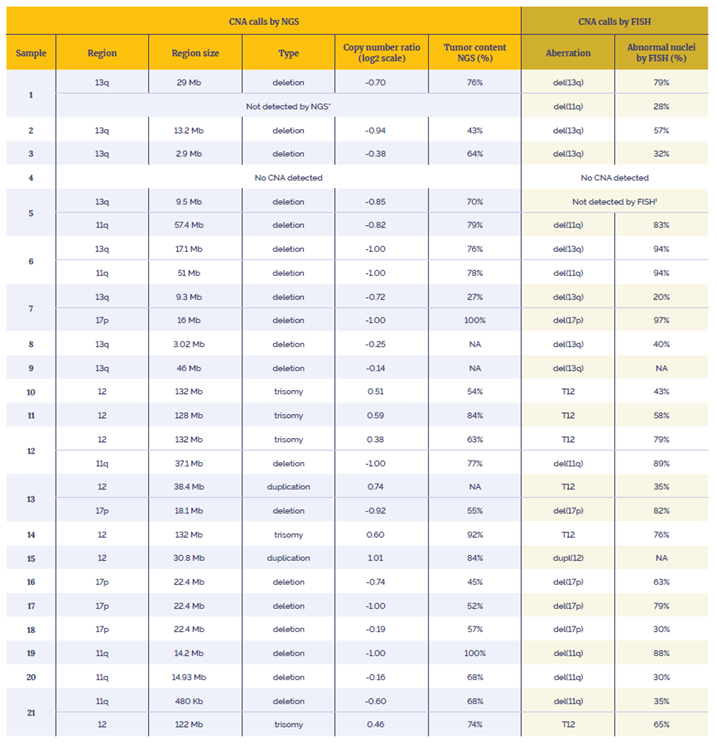 Table 3: Data generated using the SureSeq CLL + CNV V3 Panel in combination with the OGT’s Universal Complete NGS Workflow and OGT’s Interpret NGS Analysis Software. *Not enough material to perform orthogonal tests. Suspected over-estimation of tumour content by FISH, likely to be below the level of detection of the assay (20%). †Not enough material to perform orthogonal tests. Highly confident variant, log2 ratio=-0.85.
Table 3: Data generated using the SureSeq CLL + CNV V3 Panel in combination with the OGT’s Universal Complete NGS Workflow and OGT’s Interpret NGS Analysis Software. *Not enough material to perform orthogonal tests. Suspected over-estimation of tumour content by FISH, likely to be below the level of detection of the assay (20%). †Not enough material to perform orthogonal tests. Highly confident variant, log2 ratio=-0.85.
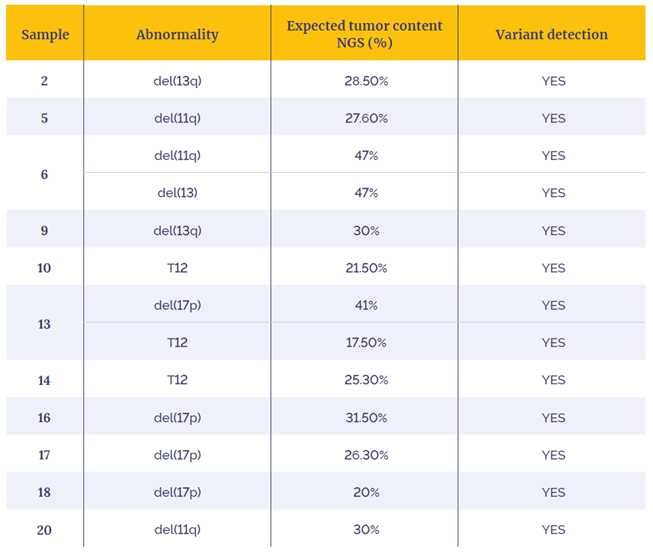 Table 4: Data generated diluting 11 research samples for low tumour content CNAs detection.
Table 4: Data generated diluting 11 research samples for low tumour content CNAs detection.
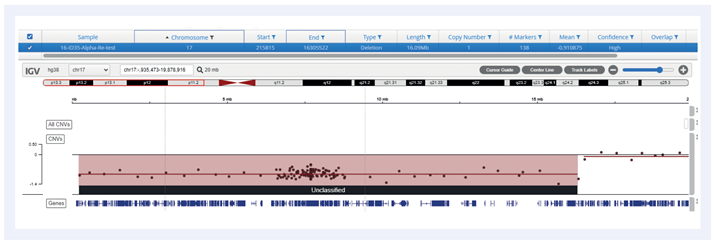 Figure 4: Example of a 16Mb deletion covering TP53 gene (del17p).
Figure 4: Example of a 16Mb deletion covering TP53 gene (del17p).
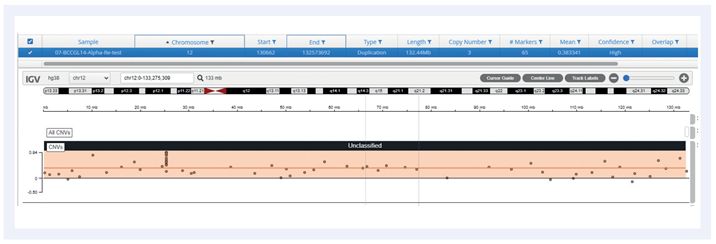
Figure 5: Example of a trisomy 12
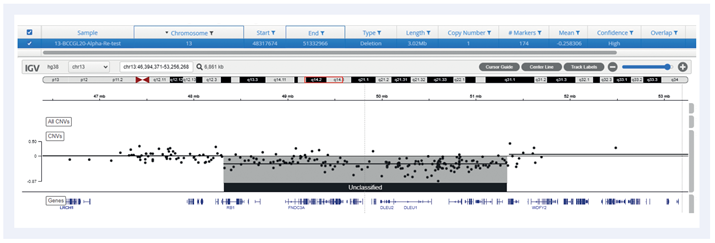
Figure 6: Example of a 3Mb deletion covering RB1 and DLEU1 genes (del13q).
SureSeq: For Research Use Only; Not for Diagnostic Procedures.
